2021 Volume 46 Issue 5 Pages 209-222
2021 Volume 46 Issue 5 Pages 209-222
Objective: To seek out the effect of curcumin on cholesterol efflux in THP-1 macrophages and clarify its specific mechanism. Methods: THP-1 macrophages were cultured with curcumin at different concentrations, followed by detection of the toxicity of curcumin to cells utilizing CCK-8 assay. Following culturing with serum-free ox-LDL, THP-1 macrophages were transfected with mi-miR-125a-5p, or in-miR-125a-5p, or pcDNA3.1-SIRT6, or si-SIRT6 for 24 hr, prior to treatment with curcumin at different concentrations. Oil red O staining was applied to examine the formation rate of foam cells, the kits were used for measuring intracellular lipid content of THP-1 macrophages, and the fluorescence detection kit for observing the cholesterol efflux rate. The expressions of miR-125a-5p, SIRT6, and ABCA1 were assayed by qRT-PCR and Western blot. ELISA was adopted to assess the contents of TNF-α, IL-6, and MCP-1. The interaction between miR-125a-5p and SIRT6 was evaluated by dual-luciferase reporter gene assay. Results: The optimal dosage of curcumin could reduce foam cell formation and intracellular lipid content, and promote cholesterol efflux in THP-1 macrophages. Meanwhile, curcumin markedly suppressed the expression of miR-125a-5p and upregulated the expression of SIRT6. MiR-125a-5p negatively targeted SIRT6. Overexpression of SIRT6 partially reversed the inhibition role of miR-125a-5p mimic in the biological function of curcumin. Silencing of SIRT6 could partially reverse the effect of the miR-125a-5p inhibitor on the biological function of curcumin. Conclusion: urcumin could promote cholesterol efflux of THP-1 macrophages through miR-125a-5p/SIRT6 axis and regulate the expression of ABCA1.
As a multifactorial chronic disease, atherosclerosis (AS) contributes to the majority of morbidity and mortality related to cardiovascular and cerebrovascular diseases (Lin et al., 2015; Yano et al., 2016). The formation of macrophage foam cells attributes to the accumulation of lipoproteins in the arterial wall that alters the structure of these lipoproteins, resulting in excessive absorption of lipoproteins by macrophages (Boshuizen et al., 2016). These lipid-loaded cells also intensify arterial wall inflammation, thus resulting in multiple fatal pathological consequences, such as rupture, hemorrhage, and calcification (Lu and Daugherty, 2015). Macrophages phagocytose cholesterol lipid droplets and also trigger cholesterol efflux, thereby reducing lipid accumulation in foam cells and delaying AS progression (Liang et al., 2019). ABCA1 presents itself as the major protein on the membrane of adipocytes, which transports cellular cholesterol and phospholipids to extracellular apoA-I by assembly of high-density lipoprotein (HDL), thereby maintaining cholesterol homeostasis (Tian et al., 2013). Thus, promoting cholesterol efflux from macrophages and reducing foam cell formation are pivotal strategies for AS prevention.
Curcumin, a traditional Chinese herb, extensively exhibits its pharmacologic application in the cardiovascular system, such as anti-oxidant and anti-inflammatory (Zhong et al., 2018). Recently, it had been found that curcumin can prevent restraint stress-induced oxidative damage in the kidney, brain, and liver (Samarghandian et al., 2017). Simultaneously, curcumin is defined to have anti-atherosclerotic effect which could increase M1 macrophages’ ability to handle harmful lipids and facilitate lipid disposal, processing, and removal (Chen et al., 2015). Suppression of cholesterol accumulation and AS through down-regulating scavenger receptor class A (SR-A) and upregulation of ABCA1 by a proteasome- and calmodulin-liver X receptor (LXR)-dependent pathway both benefits from curcumin (Zhao et al., 2012). Furthermore, epigenetic modulators such as microRNAs (miRNAs) are defined as novel targets of curcumin in renal, brain, ocular, and liver diseases (Momtazi et al., 2016). However, the molecular mechanism of their connection remains to be elucidated from different perspectives.
MiRNAs are critical regulators of numerous physiologic functions, including regulation of lipid metabolism (Price et al., 2019). An earlier study illustrated that miR-125a-5p may be involved in inflammatory response and development of AS (Wang et al., 2019). Over the past decade, increasing evidence has indicated that SIRTs could be an attractive therapeutic target in the precaution of atherosclerotic cardiovascular disease (Sosnowska et al., 2017). The importance of SIRT6 in modulating lipid metabolism also has been well-characterized. For example, SIRT6 has been reported to reduce macrophage foam cell formation by inducing autophagy and cholesterol efflux under ox-LDL condition (He et al., 2017). Additionally, SIRT6 can hinder the interaction between endothelial cells and macrophages so as to reduce the infiltration of macrophages and foam cells (Wang et al., 2020).
Given the unresolved mechanism of curcumin in managing cholesterol efflux and the implication of miR-125a-5p and SIRT6 in the setting of AS, we explored the effect of curcumin on cholesterol homeostasis of macrophages with the aim to clarify the possible mechanism herein. Curcumin can effectively enhance cholesterol efflux by mediating ABCA1 expression via the miR-125a-5p/SIRT6 signal axis in THP-1 macrophages.
Human THP-1 cells (Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences) were cultured in RPMI-1640 medium (Gibco, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS) and 1% P/S in a humidified incubator of 5% CO2 at 37°C. The culture medium was refreshed every 3 days, and the morphological feature of cells was observed under a microscope. Human THP-1 macrophages, mononuclear cells of peripheral blood, are derived from patients with acute monocytic leukemia. THP-1 macrophage belongs to suspension cells, especially sensitive to cell concentration. The optimum cell culture concentration was between 5 × 105~106/mL. The mononuclear cells were stimulated by 160 nmol/L phorbol-12-myristate-13-acetate (PMA, Promega, Beijing, China) for 24 hr to differentiate into macrophages. Then the macrophages were cultured with serum-free medium containing ox-LDL (50 mg/mL, Oxdized low density lipoprotein, Shanghai Luwen Biological Technology Co., Ltd., Shanghai, China) for 48 hr to induce foam cell formation. The hallmark of foam cells is ≥ 50% intracellular cholesterol ester (CE) and total cholesterol (TC) (Fogelman et al., 1980).
Preparation of curcumin solutionAn appropriate amount of curcumin powder (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) was weighed by microelectronic balance and dissolved in dimethylsulfoxide (DMSO) solution. The solution was prepared to a concentration of 50M and stored at -20°C. Curcumin was diluted to required concentration (0, 5, 10, 20, 40, or 80 μM) for cell treatment. The concentration of DMSO in the medium shall not exceed 0.1%.
Macrophage transfection and groupingFollowing pre-treatment with ox-LDL or without ox-LDL for 12 hr, THP-1 macrophages in the logarithmic growth phase were transfected with mimic NC, miR-125a-5p mimic (mi-miR-125a-5p), inhibitor NC, miR-125a-5p inhibitor (in-miR-125a-5p) (purchased from RiboBio, Guangzhou, China), pcDNA3.1, pcDNA3.1-SIRT6, si-NC, or si-SIRT6 (designed and synthesized by GenePharma, Shanghai, China). About 12 hr later, macrophages were treated with different concentrations (0, 5, 10, 20, 40, or 80 μM) of curcumin (Sigma-Aldrich). Cells were correspondingly grouped into the Blank group, the ox-LDL group, the ox-LDL + 5-Cur group, the ox-LDL + 10-Cur group, the ox-LDL + 20-Cur group, the ox-LDL + 40-Cur group, the ox-LDL + 80-Cur group, the ox-LDL+ 40-Cur + mimic NC group, the ox-LDL + 40-Cur + mi-miR-125a-5p group, the ox-LDL + 40-Cur + inhibitor NC group, the ox-LDL + 40-Cur + in-miR-125a-5p group, the ox-LDL + 40-Cur + pcDNA3.1 group, the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 group, the ox-LDL + 40-Cur + si-NC group, the ox-LDL + 40-Cur + si-SIRT6 group, the ox-LDL + pcDNA3.1 group, the ox-LDL + pcDNA3.1-SIRT6 group, ox-LDL + si-NC group, the ox-LDL + si-SIRT6 group, the ox-LDL + 40-Cur + pcDNA3.1 + mi-miR-125a-5p group, the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 + mi-miR-125a-5p group, the ox-LDL + pcDNA3.1 + mi-miR-125a-5p group, and the ox-LDL + pcDNA3.1-SIRT6 + mi-miR-125a-5p group. Transfection was strictly accomplished following the instructions of the Lipofectamine 2000 transfection kit (Invitrogen, Carlsbad, CA, USA). The transfected cells were added into serum-free RPMI-1640 medium for incubation at 37°C with 5% CO2 and 95% humidity. After 8 hr, the medium was replaced with the RPMI-1640 medium containing 10% FBS.
CCK-8 assayThe THP-1 macrophages in the logarithmic growth phase were mixed with 160 nmol/L PMA medium to obtain cell suspension with the cell concentration of 1 × 105 cells/FBS. The suspension was seeded in 96-well plates at the density of 10,000 cells/well and incubated at 37°C with 5% CO2 and 95% humidity for 48 hr. Following the instructions of the CCK-8 kit (Dojindo Molecular Technologies, Inc., Kumamoto, Japan), the cells in plates were cultured with RPIM-1640 medium containing different concentrations of curcumin for 24 hr, followed by adding 10 µL of CCK-8 solution into each well for incubation for 4 hr. The absorbance at the wavelength of 490 nm was measured using a microplate reader. Five wells were set up for each concentration of cells, and the absorbance of each group was measured three times and averaged.
Oil red O staining and foam cell formation rateThe THP-1 macrophages were seeded into sterile slides in the six-well plates at the density of 106 cells/well and cultured under different grouping conditions. Following 5 min of phosphate-buffered saline (PBS) washing thrice, the slides were fixed in 4% formaldehyde for 30 min, followed by Oil red O dye staining (Sigma-Aldrich) for 10 min. The slides were washed three times with PBS for 1~2 min, and counterstained with hematoxylin (Sigma-Aldrich) for 5 min to dye the nuclei and the lipid droplets. The slides were differentiated and incubated with ammonium hydroxide, followed by being mounted in glycerol solution. Images of cells were captured using a light microscope. More than intracellular 5 large red-stained particles in the cell were used as the standard for foam cells, and 10 visual fields were randomly observed in the microscope to count the total number of cells and the number of foam cells. Foam cell formation rate = foam cell number/total number of cells × 100%.
Determination of intracellular lipid contentCells were made into suspension which was centrifuged at 1,000 rpm/min for 10 min, and the supernatant was abandoned. After re-suspended in PBS, the suspension was centrifuged at 1,000 rpm/min for 10 min. The supernatant was discarded to obtain the cell sedimentation. The cell sedimentation was cultured with lysis buffer and flapped, then centrifuged at 12,000 rmp/min for 5 min to harvest the supernatant. The content of TC was tested by microplate reader (OD510) with TC assay kit (Nanjing Jiancheng Bioengineering Institute), and the content of free cholesterol (FC) was measured using TC assay kit without cholesterol esterase (CE). The content of CE = the content TC – the content of FC. The above operation was repeated to inspect the content of three acids glyceride (TG) by microplate reader (OD510) with TG assay kit (Nanjing Jiancheng Bioengineering Institute).
Measurement of cholesterol efflux rateCholesterol efflux fluorescence kit (BioVision, San Francisco, CA, USA) was applied to examine the cholesterol efflux rate. Before detection, 2 μL/mL of reagent A and reagent B in this kit were added to the equilibrium buffer solution to harvest the equilibrium medium. The THP-1 macrophages were seeded in 96-well plates at the density of 1 × 105 cells/well to obtain foam cells. The plates were washed twice with the serum-free RPMI-1640 medium, cultured with 50 μL labeling reagents and 50 μL equilibrium medium or 100 μL equilibrium medium (blank control), and incubated overnight at 37°C with 5% CO2. Cell supernatants were removed and the plates were washed with 200 μL of serum-free RPMI-1640 medium for twice. Positive control group (titrating the serum-free RPMI-1640 medium to 100 μL with 20 μL of positive control solution) and the blank control group (treated with 100 μL of RPMI-1640 medium) were set. After 6 hr incubation, the supernatant was inoculated into 96-well plates, and the fluorescence value (Ex/Em = 482/515) of the supernatant was evaluated using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). The THP-1 macrophages were treated with 10 μL of lysis buffer, and shacked for 30 min at room temperature. Then cell lysates were collected and flapped with pipette to examine the fluorescence value (Ex/Em = 482/515). The cholesterol efflux rate was calculated according to the following formula: The cholesterol efflux rate (%) = the fluorescence value of the supernatant / (the fluorescence value of the supernatant + the fluorescence value of the cell lysate) × 100%.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)The THP-1macrophages were dissolved in 1 mL of TRIzol (Invitrogen), and total RNA was extracted according to the specification. The cDNA template was synthesized by reverse transcription reaction using fluorescence quantitative PCR (Takara, Dalian, China) kit, and the qRT-PCR experiment was carried out with ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The reaction conditions were 95°C pre-denaturation for 10 min, followed by 40 cycles of 95°C denaturation for 10 sec, 60°C annealing for 20 sec, and 72°C extension for 34 sec. Then the expression levels of miR-125a-5p, SIRT6, and ABCA1 were analyzed. The internal reference of miRNA is U6 and internal reference of mRNA is GAPDH. Data analysis employs 2-ΔΔCt method (Burja et al., 2019). The formula is as follows: ΔΔCt = [Ct(target gene)-Ct(reference gene)]experimental group-[Ct(target gene)-Ct(reference gene)] control group. All primers were synthesized by Genewiz Biotechnology Co., Ltd., (GENEWIZ, Jiangsu, China) and the amplified primer sequences of each gene and its primers are shown in Table 1.
| Name of primer | Sequences |
|---|---|
| miR-125a-5p-F | GGTCATTCCCTGAGACCCTTTAAC |
| miR-125a-5p-R | GTGCAGGGTCCGAGGT |
| SIRT6-F | GCACCGTGGCTAAGGCAAGG |
| SIRT6-R | GTGATGGACAGGTCGGCGTTC |
| ABCA1-F | AGGAAACCCAATCCCAGATACCC |
| ABCA1-R | GCTCGGAGGAAGTGCTTGAGAAT |
| GAPDH-F | GCAAGGATGCTGGCGTAATG |
| GAPDH-R | TACGCGTAGGGGTTTGACAC |
| U6-F | CTCGCTTCGGCAGCACA |
| U6-R | ACGCTTCACGAATTTGCGT |
Notes: F: forward primer; R: reverse primer.
THP-1 macrophages were washed thrice with pre-cooled PBS buffer, followed by being lysed with 100 μL/50 mL RIPA lysis buffer and placed on ice for 30 min. The THP-1 cells were centrifuged at 12,000 rpm at 4°C for 10 min to acquire the supernatant, and then the supernatant was placed in a 0.5 mL centrifuge tube and stored at -20°C or quantification using a BCA kit (Beyotime, Shanghai, China). The protein was treated with 6 × SDS loading buffer for denaturation, followed by SDS electrophoresis and 90 min of PVDF membrane transfer employing 4°C pre-cooled transfer buffer. After blocked in 5% non-fat dry milk-TBST for 1 hr, the membranes were incubated with primary antibodies against SIRT6 (ab62739, 1:200, Abcam, Cambridge, MA, USA), ABCA1 (ab18180, 1:200) and β-actin (ab4970s, 1:1000) (Abcam) overnight at 4°C. After washing three times with TBST for 10 min, the membranes were incubated with goat anti-rabbit IgG or goat anti-mouse IgG (1:5000, Beijing ComWin Biotech Co., Ltd., Beijing, China) for 2 hr. The protein expression level was detected after TBST washing.
ELISAThe concentrations of TNF-α, IL-6, and MCP-1 in the supernatant of THP-1 macrophages were evaluated by ELISA kit (R&D, Minneapolis, MN, USA). All operations were performed in strict accordance with the ELISA kit instructions.
Dual-luciferase reporter gene assayThe online prediction software Starbase was employed to predict the bind site of miR-125a-5p and SIRT6. The mutated type (MT) sequences and wild type (WT) sequences in the binding sites of miR-125a-5p and SIRT6 were designed according to the predicted results and cloned into Promega vector subsequently renamed MT-SIRT6 and WT-SIRT6. Then MT-SIRT6 or WT-SIRT6 was co-transfected with miR-125a-5p mimic or miR-125a-5p inhibitor, respectively, into HEK-293T cells. Forty-eight hr after transfection, the fluorescence intensity of cells was inspected using a dual-luciferase reporter gene kit (Promega, Madison, WI, USA).
Statistical analysisData were analyzed utilizing SPSS 18.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA, USA). Data were presented as mean ± standard deviation 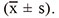 T-test was applied to the comparison between two groups, and one-way analysis of variance (ANOVA) was used for comparison among multiple groups. P < 0.05 was considered statistically significant.
T-test was applied to the comparison between two groups, and one-way analysis of variance (ANOVA) was used for comparison among multiple groups. P < 0.05 was considered statistically significant.
The THP-1 macrophages were respectively incubated with 0, 5, 10, 20, 40, and 80 μM of curcumin for 24 hr before the toxicity of curcumin was assessed by CCK-8 assay. The results presented that curcumin at the concentration of 0~40 μM showed no significant difference in the activity of macrophages (P > 0.05), while 80 μM of curcumin can markedly decreased the activity of THP-1 macrophages (Fig. 1A, P < 0.05). The ox-LDL usually applied to induce lipid absorption for detection of cholesterol efflux (Fogelman et al., 1980). In this study, macrophages were firstly stimulated by 50 μg/mL of ox-LDL, and then treated with curcumin at the concentration of 0~40 μM for 24 hr before Oil red O staining and detection of TC, FC and TG.
Curcumin enhances cholesterol efflux in THP-1 macrophages. Notes: THP-1 macrophages were incubated with 0, 5, 10, 20, 40, and 80 μM of curcumin. CCK-8 was applied to estimate the toxicity of curcumin on cell viability of macrophages (A). THP-1 macrophages were stimulated by 50 μg/mL ox-LDL and then treated with curcumin (0 ~ 40 μM) for 24 hr. The function of curcumin in foam cell formation was determined by Oil red O staining (B). Then the fluorescence detection kit was adopted to describe the role of concentration (C) and treatment time (G) of curcumin in cholesterol efflux rate. Next, the mRNA (D) and protein (E) expressions of ABCA1 were inspected by qRT-PCR and Western blot. ELISA assay was used to test the contents of TNF-α, IL-6, and MCP-1 in macrophages (F). **P < 0.01, ***P < 0.001 vs. the Blank group, #P < 0.05, ##P < 0.01 vs. the ox-LDL group.
Determination of intracellular lipid content presented that there were elevated foam cell formation, intracellular lipid content, and CE/TC value (Table 2) in the ox-LDL group (Fig. 1A, P < 0.05, vs. the Blank group). The ox-LDL + 10-Cur group, the ox-LDL + 20-Cur group, and the ox-LDL + 40-Cur group had increased cholesterol efflux rate (Fig. 1C, P < 0.05) and decreased foam cell formation (Fig. 1B, P < 0.05) as well as reduced accumulation of cholesterol and triglyceride in THP-1 macrophages (Table 2) in comparison to the ox-LDL group. There was no significant difference for comparison between the ox-LDL + 5-Cur group and ox-LDL group. These findings implicated that curcumin promotes cholesterol efflux in THP-1 macrophages in a dose-dependent manner.
| Groups | TC/mmol g-1 | FC/mmol g-1 | CE/mmol g-1 | TG/mmol g-1 | CE:TC/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blank | 5.82 | ± | 0.13 | 3.67 | ± | 0.11 | 2.16 | ± | 0.21 | 4.12 | ± | 0.12 | 37.42 | ± | 1.32 |
| ox-LDL | 20.31 | ± | 0.23** | 6.80 | ± | 0.07** | 13.01 | ± | 0.32** | 22.15 | ± | 0.42** | 64.42 | ± | 1.41** |
| ox-LDL + 5-Cur | 20.25 | ± | 0.17 | 6.73 | ± | 0.13 | 12.82 | ± | 0.13 | 21.65 | ± | 0.23 | 63.49 | ± | 1.76 |
| ox-LDL + 10-Cur | 17.05 | ± | 0.15# | 5.71 | ± | 0.13# | 9.87 | ± | 0.17# | 18.51 | ± | 0.14# | 57.96 | ± | 1.59# |
| ox-LDL + 20-Cur | 15.13 | ± | 0.21# | 5.38 | ± | 0.12# | 7.13 | ± | 0.27# | 15.27 | ± | 0.27# | 47.43 | ± | 1.42# |
| ox-LDL + 40-Cur | 13.34 | ± | 0.17## | 4.91 | ± | 0.14## | 5.72 | ± | 0.15## | 12.15 | ± | 0.21## | 43.43 | ± | 1.17## |
Notes: ox-LDL, oxidized low-density lipoprotein; Cur, curcumin; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; TG, triglyceride; *P < 0.05, **P < 0.01, vs. the Blank group; #P < 0.05, ##P < 0.01, vs. the ox-LDL group.
ABCA1 presented itself as a protein closely related to cholesterol efflux and involved in the reverse transport of cholesterol (Iizuka et al., 2012). Western blot and qRT-PCR results described that there were gradually increased mRNA and protein expressions of ABCA1 in the Blank group, the ox-LDL + 10-Cur group, the ox-LDL + 20-Cur group, and the ox-LDL + 40-Cur group (Fig. 1D-E, P < 0.05, vs. the ox-LDL group). But the expression of ABCA1 in the ox-LDL + 5-Cur group remained unchanged when compared with ox-LDL group (P > 0.05), indicating that curcumin could upregulate the expression of ABCA1.
Pro-inflammatory cytokines and inflammatory biomarkers TNF-α, IL-6, and MCP-1 are found to be elevated in AS lesions, which exacerbates insulin resistance and lipid metabolic disorder, consequently aggravates the progression of AS (Singh et al., 2005). ELISA manifested that there were suppressed contents of TNF-α, IL-6, and MCP-1 in the Blank group, the ox-LDL + 10-Cur group, the ox-LDL + 20-Cur group, and the ox-LDL + 40-Cur group (Fig. 1F, P < 0.05, vs. the ox-LDL group). The expressions of TNF-α, IL-6, and MCP-1 between ox-LDL + 5-Cur group and ox-LDL group showed no significant difference (P > 0.05). Those results showed that curcumin may reduce the contents of TNF-α, MCP-1, and IL-6 in ox-LDL-induced macrophages.
Subsequently, 40 μM curcumin was employed to stimulate THP-1 macrophages to explore the effect of curcumin stimulation time on the cholesterol efflux rate of THP-1 macrophages. The findings displayed that curcumin stimulation for 6~24 hr could continuously elevate cholesterol efflux rate and the peak cholesterol efflux rate was on the 24th hr. There was no obvious difference in cholesterol efflux rate between the 24th hr and the 48th hr (Fig. 1G, P > 0.05). In order to ensure the validity of our tests and to minimize the experimental error, THP-1 macrophages were incubated with 40 μM of curcumin for 24 hr in our later experiments.
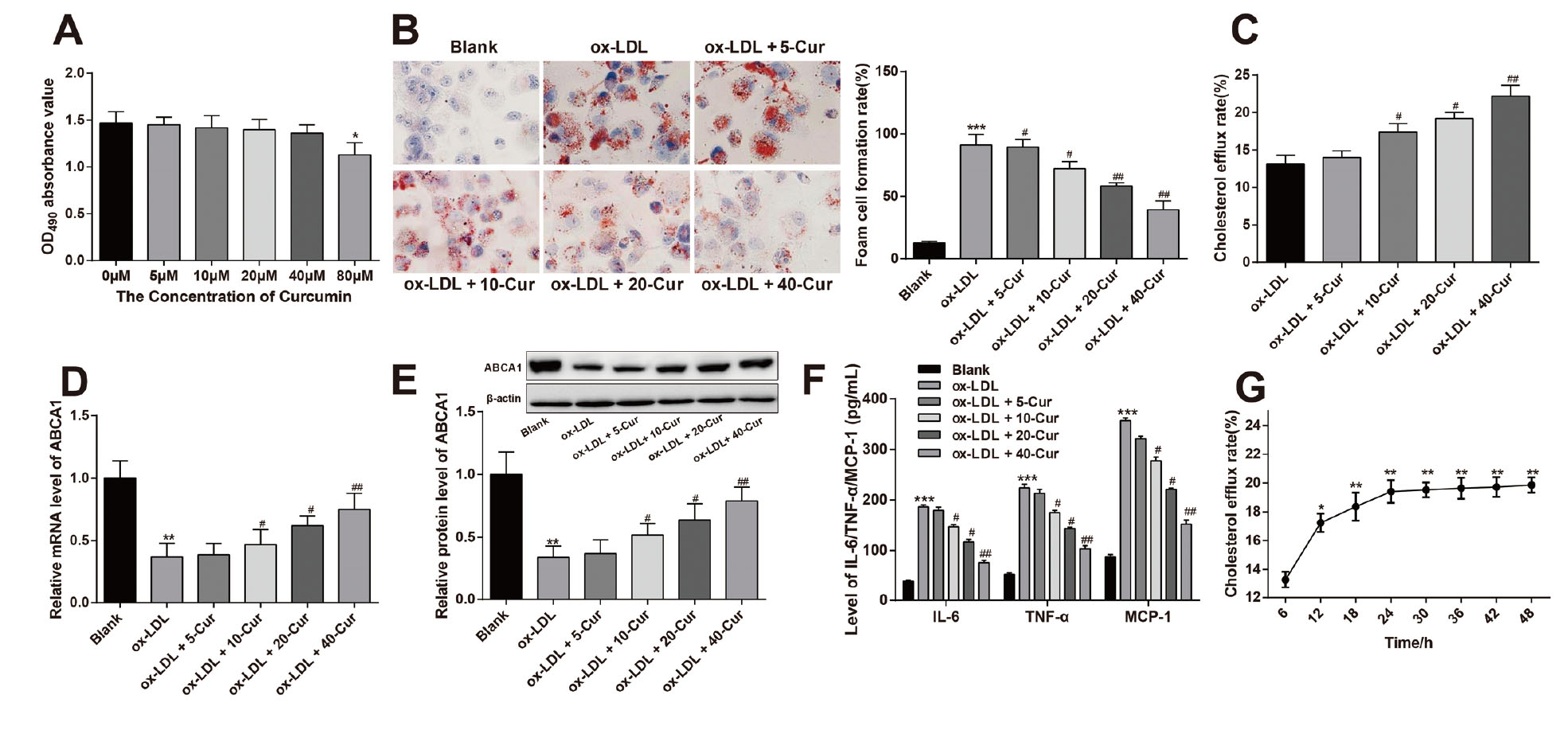
Curcumin inhibits miR-125a-5p expression and upregulates SIRT6 expression in THP-1 macrophagesTo clarify the specific mechanism of curcumin on cholesterol efflux in THP-1 macrophages, we further determined the expressions of miR-125a-5p and SIRT6 in curcumin-treated macrophages. The findings manifested that the Blank group and the ox-LDL + 40-Cur group had suppressed mRNA expression of miR-125a-5p (Fig. 2A, P < 0.01, vs. the ox-LDL group), implicating that curcumin could significantly downregulate the expression level of miR-125a-5p in ox-LDL-induced THP-1 macrophages.
The expressions of miR-125a-5p and SIRT6 can be altered by curcumin in THP-1 macrophages. Note: THP-1 macrophages were stimulated with 50 μg/mL of ox-LDL and then treated with 40 μM of curcumin. qRT-PCR and Western blot were used to test the mRNA and protein expression levels of miR-125a-5p (A) and SIRT6 (B-C). **P < 0.01, ***P < 0.001 vs. the Blank group, ##P < 0.01 vs. ox-LDL group.
A previous study reported that SIRT6 could inhibit foam cell formation under the stimulation of ox-LDL through enhancing cell autophagy and cholesterol efflux (He et al., 2017). In this research, Western blot and qRT-PCR highlighted that the Blank group and the ox-LDL + 40-Cur group had elevated mRNA and protein expression levels of SIRT6 (Fig. 2B-C, P < 0.01, vs. the ox-LDL group), expounding that curcumin can upregulate SIRT6 expression in macrophages. Taken together, curcumin promotes cholesterol efflux in THP-1 macrophages by mediating expressions of miR-125a-5p and SIRT6.
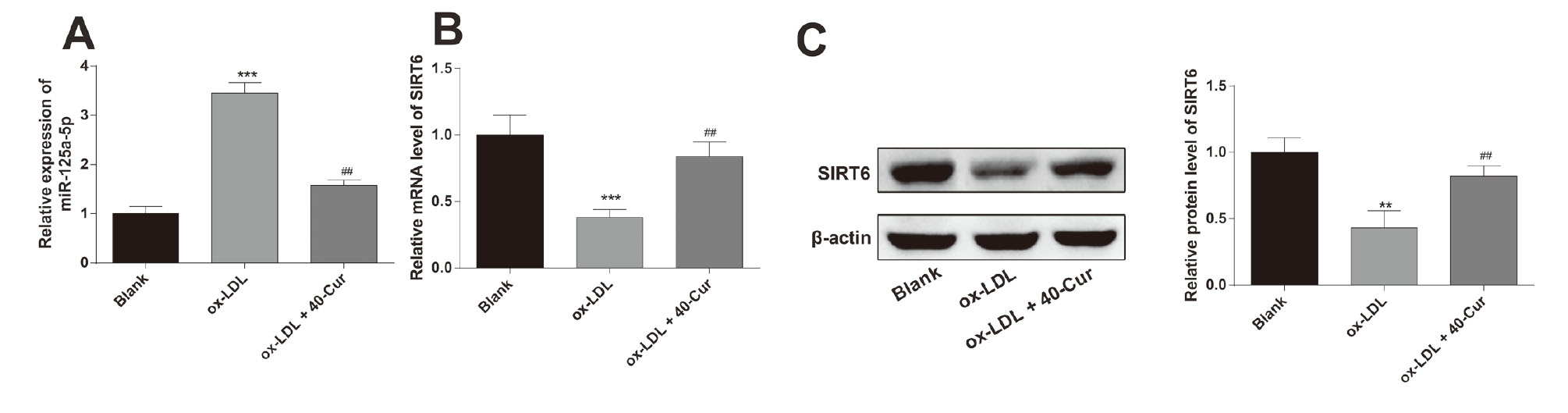
MiR-125a-5p inhibits the promotion of curcumin on cholesterol efflux in THP-1 macrophagesTo confirm the regulatory role of curcumin in miR-125a-5p in macrophages, the ox-LDL-treated THP-1 macrophages were transfected with miR-125a-5p mimic or miR-125a-5p inhibitor prior to curcumin stimulation. The qRT-PCR results displayed high expression of miR-125a-5p in the ox-LDL + 40-Cur + mi-miR-125a-5p group (Fig. 3A, P < 0.001, vs. the ox-LDL + 40-Cur + mimic NC group) and the ox-LDL + mi-miR-125a-5p group (P < 0.01, vs. the ox-LDL + mimic NC group), and low expression of miR-125a-5p in the ox-LDL + 40-Cur + in-miR-125a-5p group (Fig. 3A, P < 0.01, vs. the ox-LDL + 40-Cur + inhibitor NC group) and the ox-LDL + in-miR-125a-5p group (P < 0.01, vs. the ox-LDL + inhibitor NC group), illustrating high transfection efficiencies of miR-125a-5p mimic and miR-125a-5p inhibitor in THP-1 macrophages. Additionally, lower expression of miR-125a-5p was noticed in the curcumin-treated group than in the group without curcumin treatment.
The promotive effect of curcumin on cholesterol efflux in THP-1 macrophages can be restricted by miR-125a-5p. Notes: THP-1 macrophages were stimulated with ox-LDL for 12 hr and transfected with miR-125a-5p mimic, miR-125a-5p inhibitor or corresponding negative controls prior to stimulation with 40 μM of curcumin. Then, the transfection efficiencies ofmiR-125a-5p mimic and miR-125a-5p inhibitor were assayed by qRT-PCR (A). Overexpression or knockdown of miR-125a-5p affects the promotive effect of curcumin on cholesterol efflux in THP-1 macrophages (B). Then, qRT-PCR and Western blot were employed to evaluate the mRNA (D) and protein (C) expressions of ABCA1. The contents of TNF-α, IL-6, and MCP-1 secreted by macrophages were measured by ELISA (E). *P < 0.05, **P < 0.01, ***P < 0.001.
Meanwhile, ox-LDL + 40-Cur + mi-miR-125a-5p group and the ox-LDL + mi-miR-125a-5p group had increased intracellular lipid content and CE/TC value (Table 3), as well as decreased cholesterol efflux rate in THP-1 macrophages (Fig. 3B, P < 0.01, vs. the ox-LDL + 40-Cur + mimic NC group/ox-LDL + mimic NC group). Reverse expression pattern was found in the ox-LDL + 40-Cur + in-miR-125a-5p group and the ox-LDL + in-miR-125a-5p group group when compared with the ox-LDL + 40-Cur + inhibitor NC group or ox-LDL + inhibitor NC group (Fig. 3B, P < 0.05). Taken together, high expression of miR-125a-5p in THP-1 macrophages could inhibit cholesterol efflux of THP-1 macrophages and diminish the promotive role of curcumin in cholesterol efflux.
| Groups | TC/mmol g-1 | FC/mmol g-1 | CE/mmol g-1 | TG/mmol g-1 | CE:TC/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ox-LDL + 40-Cur + mimic NC | 14.26 | ± | 0.13 | 4.97 | ± | 0.18 | 5.89 | ± | 0.14 | 12.72 | ± | 0.15 | 42.98 | ± | 1.37 |
| ox-LDL + 40-Cur + mi-miR-125a-5p | 18.19 | ± | 0.15* | 5.89 | ± | 0.12* | 9.24 | ± | 0.17** | 19.11 | ± | 0.13** | 51.79 | ± | 1.63** |
| ox-LDL + 40-Cur + inhibitor NC | 14.39 | ± | 0.12 | 4.95 | ± | 0.15 | 5.81 | ± | 0.13 | 12.19 | ± | 0.21 | 42.15 | ± | 1.18 |
| ox-LDL + 40-Cur + in-miR-125a-5p | 9.96 | ± | 0.13## | 4.21 | ± | 0.12# | 3.91 | ± | 0.18# | 7.94 | ± | 0.16# | 37.42 | ± | 1.57# |
| ox-LDL + mimic NC | 21.48 | ± | 0.16 | 6.92 | ± | 0.16 | 13.17 | ± | 0.15 | 22.43 | ± | 0.17 | 61.74 | ± | 1.16 |
| ox-LDL + mi-miR-125a-5p | 25.14 | ± | 0.11& | 7.43 | ± | 0.13& | 17.46 | ± | 0.18& | 25.89 | ± | 0.14& | 68.59 | ± | 1.72& |
| ox-LDL + inhibitor NC | 21.05 | ± | 0.14 | 6.78 | ± | 0.14 | 13.49 | ± | 0.11 | 21.98 | ± | 0.18 | 60.85 | ± | 1.68 |
| ox-LDL + in-miR-125a-5p | 16.83 | ± | 0.13@@ | 5.83 | ± | 0.18@ | 9.18 | ± | 0.15@ | 16.73 | ± | 0.15@ | 49.48 | ± | 1.49@@ |
Notes: ox-LDL, oxidized low-density lipoprotein; Cur, curcumin; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; TG, triglyceride; *P < 0.05, **P < 0.01, vs. the ox-LDL + 40-Cur + mimic NC group; #P < 0.05, ##P < 0.01, vs. the ox-LDL + 40-Cur + inhibitor NC group; &P < 0.05, vs. the ox-LDL + mimic NC group; @P < 0.05, @@P < 0.01, vs. the ox-LDL + inhibitor NC group.
The qRT-PCR, Western blot, and ELISA results manifested that there were elevated contents of IL-6, TNF-α, and MCP-1 and suppressed mRNA and protein expression levels of ABCA1 in the ox-LDL + 40-Cur + mi-miR-125a-5p group and the ox-LDL + mi-miR-125a-5p group (Fig. 3C-E, P < 0.01, vs. the ox-LDL + 40-Cur + mimic NC group/ox-LDL + mimic NC group). However, these trends were totally reversed in the ox-LDL + 40-Cur + in-miR-125a-5p group and the ox-LDL + in-miR-125a-5p group (Fig. 3C-E, P < 0.05, vs. the ox-LDL + 40-Cur + inhibitor NC group/ox-LDL + inhibitor NC group). Taken together, miR-125a-5p mimic inhibits ABCA1 expression and promotes the secretion of IL-6, TNF-α and MCP-1 in macrophages, while miR-125a-5p inhibitor enhances the expression of ABCA1 and inhibits inflammatory response of macrophages. Furthermore, treatment with curcumin can potentiate ABCA1 expression and suppress the secretion of inflammatory biomarkers in macrophages.
SIRT6 enhances the regulation of curcumin on cholesterol efflux in THP-1 macrophagesTo analyze whether SIRT6 affects the role of curcumin in regulating cholesterol efflux of THP-1 macrophages, THP-1 macrophages were stimulated with 50 μg/mL of ox-LDL for 12 hr and then transfected with pcDNA3.1-SIRT6, si-SIRT6 or corresponding negative controls before stimulation with 40 μM of curcumin. Analyses of qRT-PCR and Western blot showed that the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 group and the ox-LDL + pcDNA3.1-SIRT6 group had increased mRNA and protein expressions of SIRT6 (Fig. 4A-B, P < 0.01, vs. the ox-LDL + 40-Cur + pcDNA3.1 group/ox-LDL + pcDNA3.1 group), and the ox-LDL + 40-Cur + si-SIRT6 group and the ox-LDL + si-SIRT6 group had suppressed SIRT6 expression (Fig. 4A-B, P < 0.01, vs. the ox-LDL + 40-Cur + si-NC group/ox-LDL + si-NC group). These observations illustrated the transfection efficiencies of pcDNA3.1-SIRT6 or si-SIRT6 in THP-1 macrophages. Additionally, exposure to curcumin notably enhanced the expression of SIRT6.

The promotive effect of curcumin on cholesterol efflux in macrophages can be enhanced by SIRT6. Notes: THP-1 macrophages were stimulated with 50 μg/mL of ox-LDL for 12 hr, and then transfected with pcDNA3.1-SIRT6, si-SIRT6 or corresponding negative controls before stimulation with 40 μM of curcumin. The detection of the mRNA (A) and protein expressions (B) of SIRT6 was conducted by qRT-PCR and Western blot. Overexpression or knockdown of SIRT6 affects the regulation of curcumin on cholesterol efflux in THP-1 macrophages (C). Then qRT-PCR and Western blot were adopted to assay the mRNA (D) and protein (E-F) expressions of ABCA1. The contents of TNF-α, IL-6, and MCP-1 secreted by macrophages were detected by ELISA (G). *P < 0.05, **P < 0.01, ***P < 0.001.
The fluorescence detection results showed that there were elevated cholesterol efflux rate, and suppressed intracellular lipid and CE/TC value (Table 4) in THP-1 macrophages in the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 group and the ox-LDL + pcDNA3.1-SIRT6 group (Fig. 4C, P < 0.01, vs. the ox-LDL + 40-Cur + pcDNA3.1 group/ox-LDL + pcDNA3.1 group). Reverse trends were noticed in the ox-LDL + 40-Cur + si-SIRT6 group and the ox-LDL + si-SIRT6 group (Fig. 4C, P < 0.05, vs. the ox-LDL + 40-Cur + si-NC group/ox-LDL + si-NC group). These results indicated that overexpression of SIRT6 could facilitate cholesterol efflux of macrophages and enhance the promotive role of curcumin in cholesterol efflux in THP-1 macrophages.
| Groups | TC/mmol g-1 | FC/mmol g-1 | CE/mmol g-1 | TG/mmol g-1 | CE:TC/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ox-LDL + 40-Cur + pcDNA3.1 | 13.68 | ± | 0.16 | 4.73 | ± | 0.13 | 5.82 | ± | 0.15 | 12.14 | ± | 0.14 | 43.85 | ± | 1.72 |
| ox-LDL + 40-Cur + pcDNA3.1-SIRT6 | 9.14 | ± | 0.13** | 3.81 | ± | 0.12* | 3.32 | ± | 0.13* | 8.19 | ± | 0.21** | 37.76 | ± | 1.59* |
| ox-LDL + 40-Cur + si-NC | 13.42 | ± | 0.17 | 4.76 | ± | 0.16 | 5.78 | ± | 0.14 | 12.17 | ± | 0.21 | 43.26 | ± | 1.19 |
| ox-LDL + 40-Cur + si-SIRT6 | 18.73 | ± | 0.11# | 5.79 | ± | 0.14# | 9.41 | ± | 0.12## | 18.23 | ± | 0.23## | 54.32 | ± | 1.87## |
| ox-LDL + pcDNA3.1 | 20.73 | ± | 0.14 | 6.83 | ± | 0.15 | 12.84 | ± | 0.13 | 22.24 | ± | 0.16 | 62.17 | ± | 2.13 |
| ox-LDL + pcDNA3.1-SIRT6 | 15.78 | ± | 0.18& | 5.24 | ± | 0.13& | 8.31 | ± | 0.17& | 17.19 | ± | 0.14&& | 49.74 | ± | 1.67&& |
| ox-LDL + si-NC | 20.52 | ± | 0.09 | 6.79 | ± | 0.17 | 12.75 | ± | 0.15 | 22.11 | ± | 0.13 | 61.62 | ± | 1.96 |
| ox-LDL + si-SIRT6 | 24.46 | ± | 0.15@ | 7.34 | ± | 0.12@ | 14.93 | ± | 0.16@ | 25.37 | ± | 0.18@ | 70.23 | ± | 2.25@@ |
Notes: ox-LDL, oxidized low-density lipoprotein; Cur, curcumin; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; TG, triglyceride; *P < 0.05, **P < 0.01, vs. the ox-LDL + 40-Cur + pcDNA3.1 group; #P < 0.05, ##P < 0.01, vs. the ox-LDL + 40-Cur + si-NC group; &P < 0.05, vs. the ox-LDL + pcDNA3.1 group; @P < 0.05, @@P < 0.01, vs. the ox-LDL + si-SIRT6 group.
The qRT-PCR, Western blot, and ELISA detection showed that the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 group and the ox-LDL + pcDNA3.1-SIRT6 group had increased mRNA and protein expression levels of ABCA1 (Fig. 4D-F, P < 0.01) and decreased contents of TNF-α, IL-6, and MCP-1 (Fig. 4G, P < 0.05) in comparison to the ox-LDL + 40-Cur + pcDNA3.1 group or ox-LDL + pcDNA3.1 group. Different expression profile of above parameters was found in the ox-LDL + 40-Cur + si-SIRT6 group and the ox-LDL + si-SIRT6 group in comparison to the ox-LDL + 40-Cur + si-NC group or ox-LDL + si-NC group (Fig. 4D-G, P < 0.05). All these findings implicated that overexpression of SIRT6 or curcumin treatment reinforces the expression of ABCA1 and decreases the secretion of TNF-α, IL-6 and MCP-1.
MiR-125a-5p negatively regulates SIRT6Based on the above experimental results, we hypothesized that miR-125a-5p may be an upstream target of SIRT6. The online prediction software Starbase was employed to predict the binding relationship between miR-125a-5p and SIRT6 (Fig. 5A). We discovered that SIRT6 expression in the miR-125a-5p mimic group was decreased markedly (Fig. 5B-D, P < 0.01, vs. the mimic NC group) and SIRT6 expression in the miR-125a-5p inhibitor group was increased notably (Fig. 5B-D, P < 0.01, vs. the inhibitor NC group). These results indicated that miR-125a-5p negatively regulates SIRT6 expression.
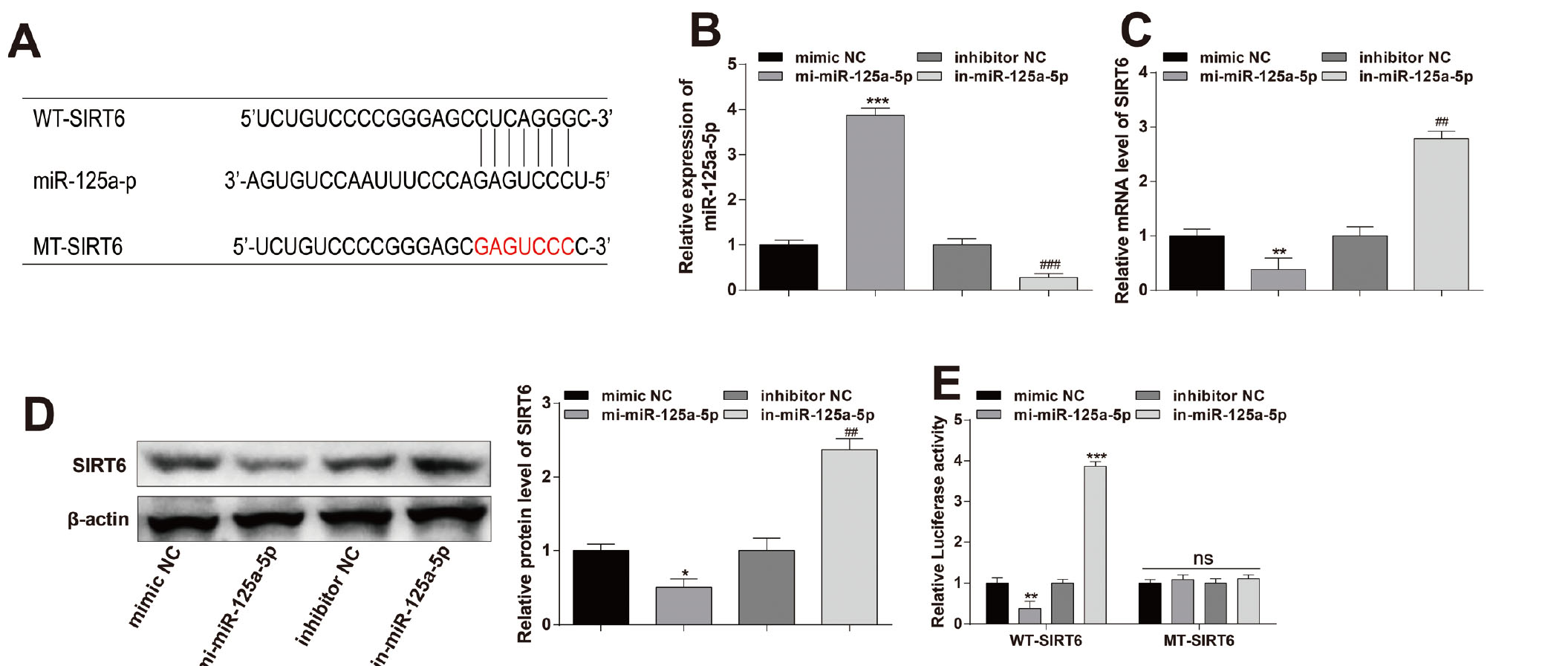
MiR-125a-5p negatively targets SIRT6. Notes: THP-1 macrophages were transfected with miR-125a-5p mimic or miR-125a-5p inhibitor or corresponding negative control. The binding site of miR-125a-5p and SIRT6 was predicted by Starbase (A). The transfection efficiency of miR-125a-5p mimic or miR-125a-5p inhibitor was determined by qRT-PCR (B). qRT-PCR and Western blot were applied to inspect the mRNA (C) and protein (D) expressions of SIRT6. After transfection or co-transfection of miR-125a-5p mimic or miR-125a-5p inhibitor or corresponding negative control with WT-SIRT6 or MT-SIRT6 into HEK-293T cells, the luciferase activity was evaluated by dual-luciferase reporter gene assay (E). *P < 0.05, **P < 0.01, ***P < 0.001 vs. the mimic NC group, ##P < 0.01, ###P < 0.001 vs. the inhibitor NC group; NC, negative control.
Subsequently, a wild-type SIRT6 luciferase promoter plasmid (named WT-SIRT6) and a mutant SIRT6 luciferase promoter plasmid (MT-SIRT6, containing 10 mutation sites predicted to bind to miR-21-5p) were constructed to unearth the targeting relationship between miR-125a-5p and SIRT6. We manifested that the luciferase activity of WT-SIRT6 was repressed when WT-SIRT6 was co-transfected with miR-125a-5p mimic in HEK-293T cells (Fig. 5E, vs. co-transfected with mimic NC). Co-transfection of WT-SIRT6 with miR-125a-5p inhibitor in HEK-293T cells had increased luciferase activity of WT-SIRT6 (Fig. 5E, vs. co-transfected with inhibitor NC). There was no significant difference when MT-SIRT6 was co-transfected with miR-125a-5p mimic or miR-125a-5p inhibitor (Fig. 5E, vs. co-transfected with mimic NC or inhibitor NC). All these findings implicated that SIRT6 directly binds to miR-125a-5p.
Curcumin promotes cholesterol efflux in THP-1 macrophages via miR-125a-5/SIRT6 axisTo verify whether curcumin promotes cholesterol efflux in THP-1 macrophage through the miR-125a-5p/SIRT6 signal axis, ox-LDL-induced THP-1 macrophages were co-transfected with mi-miR-125a-5p or in-miR-125a-5p and pcDNA3.1-SIRT6 or si-SIRT6. Then THP-1 macrophages were stimulated with 40 μM of curcumin or did nothing.
We presented that the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 + mi-miR-125a-5p group and the ox-LDL + pcDNA3.1-SIRT6 + mi-miR-125a-5p group had elevated cholesterol efflux rate and suppressed intracellular lipid and CE/TC value (Table 5) in THP-1 macrophages (Fig. 6A, P < 0.05, vs. ox-LDL + 40-Cur + pcDNA3.1 + mi-miR-125a-5p group/ox-LDL + pcDNA3.1 + mi-miR-125a-5p group). These results explained that curcumin can facilitate cholesterol efflux from THP-1 macrophages, overexpression of miR-125a-5p can inhibit the promotive role of curcumin in cholesterol efflux from THP-1 macrophages, while overexpression of SIRT6 can partially reverse the inhibitory effect of miR-125a-5p mimic.
| Groups | TC/mmol g-1 | FC/mmol g-1 | CE/mmol g-1 | TG/mmol g-1 | CE:TC/% | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ox-LDL + 40-Cur + pcDNA3.1 + mi-miR-125a-5p | 17.43 | ± | 0.13 | 5.89 | ± | 0.14 | 9.29 | ± | 0.15 | 18.61 | ± | 0.19 | 52.66 | ± | 1.97 |
| ox-LDL + 40-Cur + pcDNA3.1-SIRT6 + mi-miR-125a-5p | 8.61 | ± | 0.16** | 4.11 | ± | 0.19* | 4.78 | ± | 0.12** | 11.87 | ± | 0.14** | 45.17 | ± | 2.32* |
| ox-LDL + pcDNA3.1 + mi-miR-125a-5p | 24.73 | ± | 0.18 | 7.52 | ± | 0.21 | 16.17 | ± | 0.13 | 25.14 | ± | 0.16 | 67.85 | ± | 1.58 |
| ox-LDL + pcDNA3.1-SIRT6 + mi-miR-125a-5p | 19.87 | ± | 0.14## | 6.37 | ± | 0.17# | 11.53 | ± | 0.21## | 20.26 | ± | 0.22## | 59.28 | ± | 1.82## |
Notes: ox-LDL, oxidized low-density lipoprotein; Cur, curcumin; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; TG, triglyceride; *P < 0.05, **P < 0.01, vs. the ox-LDL + 40-Cur + pcDNA3.1 + mi-miR-125a-5p group; #P < 0.05, ##P < 0.01, vs. the ox-LDL + pcDNA3.1 + mi-miR-125a-5p group.
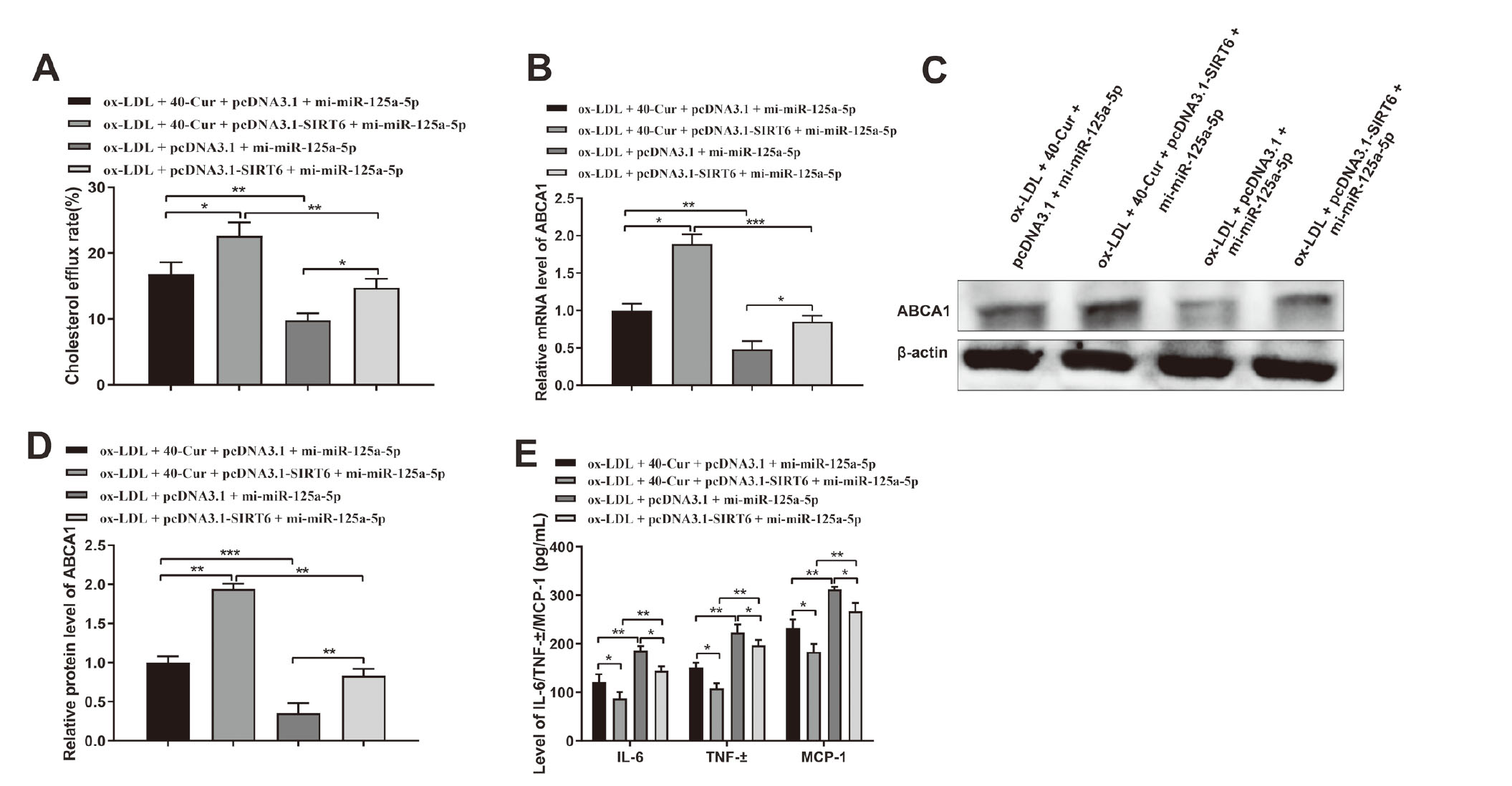
miR-125a-5p/SIRT6 axis is involved in curcumin enhancing cholesterol efflux in THP-1 macrophages. Notes: After stimulation with 50 μg/mL ox-LDL for 12 hr, THP-1 macrophages were co-transfected with mi-miR-125a-5p, or in-miR-125a-5p and pcDNA3.1-SIRT6, or si-SIRT6 prior to treatment with 40 μM of curcumin. The cholesterol efflux fluorescence detection kit was employed to determine cholesterol efflux rate (A). Then qRT-PCR and Western blot were adopted to assay the mRNA (B) and protein (C-D) expressions of ABCA1. The contents of TNF-α, IL-6, and MCP-1 secreted by macrophages were determined by ELISA (E). *P < 0.05, **P < 0.01, ***P < 0.001.
Compared with the ox-LDL + 40-Cur + pcDNA3.1 + mi-miR-125a-5p group or the ox-LDL + pcDNA3.1 + mi-miR-125a-5p group, the ox-LDL + 40-Cur + pcDNA3.1-SIRT6 + mi-miR-125a-5p group or the ox-LDL + pcDNA3.1-SIRT6 + mi-miR-125a-5p group had higher mRNA and protein expressions of ABCA1 (Fig. 6B-D, P < 0.05) and lower contents of TNF-α, IL-6, and MCP-1 (Fig. 6E, P < 0.05). All these finding ascertained that overexpression of SIRT6 could reverse the inhibitory effect of miR-125a-5p mimic on ABCA1 expression in macrophages, and inhibit the secretion of TNF-α, MCP-1, and IL-6. Curcumin treatment can enhance ABCA1 and repress the secretion of TNF-α, MCP-1, and IL-6. Taken together, the above results illustrated that curcumin promotes cholesterol efflux in THP-1 macrophages via the miR-125a-5p/SIRT6 signal axis.
The inflammatory response initiated by lipid-laden macrophages and the regulation of cholesterol metabolism is inevitable to the initiation and progression of AS (Lien et al., 2019). An increased macrophage cholesterol efflux was proven to significantly and inversely associate with cardiovascular diseases (Wang et al., 2018). Herein, THP-1 macrophages were stimulated to induce foam cell formation to test the potential functions and possible mechanism of curcumin in cholesterol efflux of macrophages. Our findings demonstrated that curcumin could enhance cholesterol efflux by upregulating ABCA1 expression through the miR-125a-5p/SITR6 axis.
The reverse cholesterol transport (RCT), particularly from macrophages, has been verified to hinder AS progression, through which the cholesterol was transported to the liver for excretion into the bile (Hafiane et al., 2019). ABCA1, a plasma membrane protein, acts as the first step in RCT and exports FC and phospholipids from cells to lipid-poor apoA-I (Kuang et al., 2017). In this study, we found curcumin could elevate the protein expression of ABCA1 in macrophages. Consistent with our findings, curcumin was reported to retune cholesterol transport homeostasis by promoting ABCA1 expression. For example, Dong et al. (2011) stated that curcumin enhances cholesterol efflux from adipocytes via the PPARc-LXR-ABCA1 pathway, by which curcumin increases mRNA level of ABCA1 by promoting PPARc-LXRa transcriptional activity. A similar standpoint declared by Lin et al. (2015) displayed that the beneficial effects of curcumin on the promotion of cholesterol efflux by enhancing ABCA1 expression through activating AMPK-SIRT1-LXRα signaling in THP-1 macrophage-derived foam cells. This evidence corroborated our findings that curcumin can dramatically enhance the expression of ABCA1 through different pathways. Although curcumin enhances cholesterol efflux in macrophages by upregulating ABCA1, the detailed mechanism of curcumin in suppressing cholesterol accumulation still needs to be further analyzed.
In AS, miR-125a-5p mediates oxLDL-induced pyroptosis in vascular endothelial cells by down-regulating TET2 (Zhaolin et al., 2019). Former study also put forward that miR-125a may inhibit innate macrophage responses by mediating macrophage inflammation, differentiation, and autophagy (Lee et al., 2016). In our study, we initially aimed to recognize whether curcumin promotes cholesterol efflux in THP-1 macrophages through regulating miR-125a-5p. Hence, THP-1 macrophages were transfected with miR-125a-5p mimic or miR-125a-5p inhibitor before curcumin treatment. Our study presented that overexpression of miR-125a-5p decreased the expression of ABCA1, and reduced the contributive effect of curcumin on cholesterol efflux in THP-1 macrophages. As expected, consistent evidence was found in cells transfected with knockdown of miR-125a-5p. These data confirmed that the alteration of miR-125a-5p can influence the effect of curcumin in cholesterol efflux.
To probe the downstream protein of miR-125a-5p in macrophages, we screened SIRT6 to seek further evidence for the potential synergistic effect of SIRT6 on the regulation of ABCA1 and cholesterol efflux. ELISA, Western blot, and qRT-PCR presented that overexpression of SIRT6 enhanced the effect of curcumin in regulating ABCA1 and cholesterol efflux, while reducing the secretion of pro-inflammatory cytokines. Similar findings were displayed in cells transfected with knockdown of SIRT6. The biological function of SIRT6 emerged in cardiovascular diseases, including cardiac hypertrophy, heart failure, myocardial hypoxic damage, and metabolism (Winnik et al., 2015). The SIRT6-mediated PKM2 pathway participates in the alleviation of inflammation of vascular endothelial cells by hydroxytyrosol acetate (Yao et al., 2019). Subsequently, SIRT6 has been revealed to reduce the formation of atherosclerotic lesions via the attenuation of endothelial dysfunction and vascular inflammation (Xu et al., 2016). More importantly, SIRT6 plays a pivotal role in the regulation of hepatic Srebp2 gene expression as well as cholesterol homeostasis (Tao et al., 2013). With regards to this evidence, we ensured that SIRT6 is an indispensable factor for curcumin to promote cholesterol efflux to achieve cholesterol homeostasis. Furthermore, SIRT6 is defined as a target gene of miR-125a-5p. Taken together, curcumin can promote cholesterol efflux in THP-1 macrophages by suppressing the expression of miR-125a-5p and upregulating the expression of SIRT6 to reinforce the expression of ABCA1.
In conclusion, evidence in this study supported the promotive effect of curcumin on cholesterol efflux in THP-1 macrophages. Curcumin could upregulate the expression of ABCA1 through miR-125a-5p/SIRT6 axis to enhance cholesterol efflux in THP-1 macrophages. Aside from miR-125a-5p, previous study proposed that other miRNAs may interfere with the effect of curcumin, such as hsa-miR-574-3p and hsa-miR-210 (Gao et al., 2014). Therefore, the regulation of curcumin on other microRNAs such as hsa-miR-574-3p and hsa-miR-210 can serve as one of the directions in our future research.
Conflict of interestThe authors declare that there is no conflict of interest.